Identification of concentration/purity and integrity of nucleic acid identification methods

Text / Han Ying Editor / Bruce.Y
Picture / Network + Original

Nucleic acid purification has been widely used in molecular biology laboratories. How to obtain high-quality, high-purity DNA or RNA samples is critical to the smooth progress of downstream experiments. Therefore, the identification of purified nucleic acids is also an essential step. This article briefly reviews the methods of nucleic acid identification.
Nucleic acid identification includes: concentration / purity identification + integrity identification
1. Concentration / purity identification
(1) Ultraviolet spectrophotometer:
Principle: Since the bases in the nucleic acid all have a conjugated double bond, they have ultraviolet light absorbing properties. The nucleic acid has a typical absorption curve in the ultraviolet region with an absorption peak at 260 nm and an absorption valley at 230 nm. The absorption peak of protein in the ultraviolet region is at 280 nm, so the ultraviolet absorption spectrum data of nucleic acid is one of the important basis for identifying nucleic acids.
Since the maximum absorption wavelength of the nucleotide is 260 nm, according to Lambert-Beer's law, the optical density of one OD value is approximately equivalent to 50 μg/ml of double-stranded DNA and 40 μg/ml of single-stranded light under ultraviolet light of 260 nm. RNA. Ultraviolet spectrophotometry is only suitable for the determination of nucleic acid solutions with concentrations greater than 0.25 μg/ml.
Calculation formula:
dsDNA (μg / μl) = OD260x50x dilution factor / 1000
RNA (μg / μl) = OD260x40x dilution factor / 1000
When OD260/OD280< 1.7, indicating high protein content, it can be extracted by phenol, chloroform, isoamyl alcohol, and then precipitated by ethanol precipitation.
When OD260/OD280> 2.0, it indicates a higher RNA content. Purification can be carried out after digestion with RNaseA.
The value of OD260/OD230 is generally not less than 2.0. If it is less than 2.0, it indicates contamination with carbohydrates, salts or organic reagents.
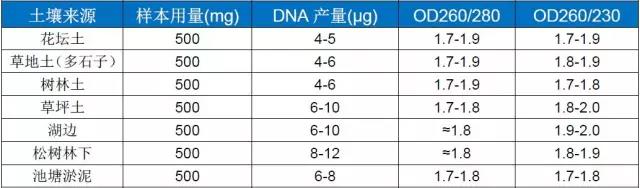
Example: Genomic DNA extraction yield and purity
FOREGENE Soil DNA Isolation Kit ( DE-05511)
Features:
Easy – no grinding required, no need for low temperature operation.
Fast - complete the operation within 90 minutes.
Safe—no organic reagents are required.
This kit handles soil samples from various sources. The amount and purity of genomic DNA obtained are shown in the following table:
Pure RNA OD260/OD280=2.0, the prepared sample DNA ratio is 1.8-2.1, OD260/OD280 ratio will be affected by the pH value of the solution used, for example, the purified RNA in the pH=7.5 buffer OD260/OD280 reading Between 1.9 and 2.1, the ratio in the neutral aqueous solution will be low, probably only 1.8-2.0, which does not mean that the quality of the RNA is deteriorated, so electrophoresis detection is also needed.
When OD260/OD280<1.8, there may be contamination of proteins or other organic substances in the solution.
The value of OD260/OD230 is generally not less than 2.0. If it is less than 2.0, it indicates the residue of strontium salt, β-mercaptoethanol or ethanol, but the ratio may vary depending on the sample. The mouse kidney and brain tissue are extracted as shown in the following table. The RNA260/230 ratio is too small.
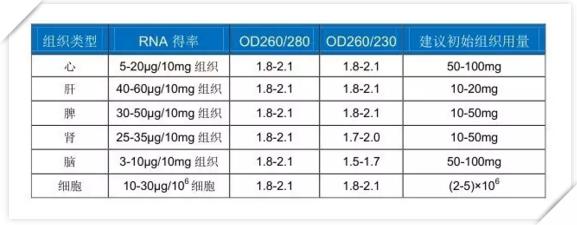
Example: Tissue RNA extraction yield and purity
FOREGENE Animal Total RNA Isolation Kit (RE-03012)
Features:
Easy – no need for low temperature operation.
Fast - complete the operation within 30 minutes.
Safe—no organic reagents are required.
This kit handles various mouse tissue samples. The amount and purity of RNA obtained are shown in the following table:
(2) Fluorescence spectrophotometry:
Fluorescence spectrophotometry, after the nucleic acid dye ethidium bromide EB is inserted into the base plane, the nucleic acid which is not fluorescent itself emits orange fluorescence under ultraviolet excitation, and the fluorescence intensity integral is proportional to the nucleic acid content. The method has a sensitivity of 1-5 ng and is suitable for quantitative analysis of low concentration nucleic acid solutions. In addition, SYBR Gold is a new ultra-sensitive fluorescent fuel that detects less than 20pg of double-stranded DNA from an agarose gel.
The purity of the nucleic acid can be determined by electrophoresis results of nucleic acid traced by a fluorescent dye such as ethidium bromide. Because DNA molecules are much larger than RNA, electrophoretic mobility is low; RNA has the most rRNA, accounting for 80-85%, tRNA and nuclear small RNA account for 15%-20%, and mRNA accounts for 1%-5%. After RNA electrophoresis, characteristic three bands can be presented. In prokaryotes, there are clearly visible 23S, 16S rRNA bands and bands composed of 5S rRNA and tRNA. Eukaryotes are 28S, 18S rRNA and a band consisting of 5S, 5.8S rRNA and tRNA. The lack of mRNA and the size of the molecules are generally invisible. By electrophoresis we can identify the presence or absence of DNA contamination in RNA products and identify the presence or absence of RNA contamination in DNA products.

2. Integrity detection
Conventionally, agarose gel electrophoresis is used. The molecular weight of genomic DNA is very large, and it is very slow to move in an electric field. If a small molecule DNA fragment is degraded, it can be significantly expressed on the electrophoresis pattern.
Complete RNA degradation map with no degradation or little degradation, except for the characteristic three bands, the fluorescence intensity of 28S (or 23S) RNA is about twice that of 18S (or 16S) RNA, otherwise it suggests RNA degradation. (figure 1).
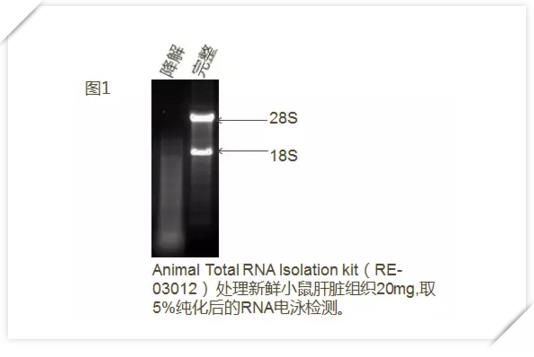
There may be more than three bands in the RNA sample electrophoresis (Figure 2). Please don't worry, because some organelles in the cell also contain RNA, such as mitochondria and chloroplasts. If there is a colored band near the electrophoresis well, DNA contamination is indicated.

RNA integrity can also be analyzed, if necessary, by special experiments, such as small-scale first-strand cDNA synthesis reactions.
Fuji Bio has developed a corresponding nucleic acid purification kit for different sample types.
FOREGENE's unique filter column separates DNA and RNA directly, and you can get high-quality nucleic acids without adding the corresponding nuclease! Convenient and fast, good quality and cheap!
Welcome new and old customers to order!

Fuji Bio Direct PCR Kit does not require separate nucleic acid purification, and the sample can be directly added to the PCR tube containing the Fuji Direct PCR System for reaction!
Fitness Equipment,Folding Fitness Equipment,Fitness Mini Gym Equipment,Home Fitness Equipment
Haloxylon Ammodendron Medical Equipment Co., Ltd. , https://www.ssmedicaldevic.com