JCI's latest publication: Application of Arraystar ChIRP sequencing and RNA sequencing in diabetic nephropathy
Professor Farhad R. Danesh of the University of Texas at Anderson Cancer Research Center is mainly engaged in the basic and clinical research of renal pathology. Recently, its laboratory found that the lncRNA molecule, Tug1, was lowly expressed in diabetic mice by Arraystar RNA-seq. The expression profile chip and Arraystar CHIRP-seq demonstrated that Tug1 can directly bind to the TBE elements of the upstream promoter regions of PGC-1α and Ppargc1α, promote the expression of Ppargc1α gene, and ultimately affect the mitochondrial bioenergy of podocytes in diabetic patients. The research results were published in the International Top Medical Journal Journal of Clinical Investigation (IF=12.81). ( RNA-seq, expression profile chip, CHIRP-seq provided by Arraystar )
Background Diabetic nephropathy (DN) is a common microvascular complication of diabetes in the United States and the leading cause of end stage renal disease. In the study of DN pathology-related factors, it was found that cofactor 1α (PGC-1α, encoded by Ppargc1α), a peroxisome proliferator-activated receptor γ, is significantly underexpressed in diabetic patients, and this factor regulates mitochondrial contents and The expression of oxidative phosphorylation gene proves to be at the core of the regulation of mitochondrial bioenergy and respiratory function. PGC-1α is in an autoregulatory cycle, and related factors involved in its regulation are associated with cellular energy and mitochondrial homeostasis. In view of the fact that PGC-1α is a key factor in linking physiological stimulation with mitochondrial bioenergy, its regulation must be rigorous and multi-layered. Some new mechanisms for regulating PGC-1α are worth exploring. The role of lncRNA in the regulation of PGC-1α has been little known. Professor Farhad R. Danesh's main research aims to reveal the evolutionarily conserved lncRNA-Tug1, regulate the expression of PGC-1α, and explore its regulatory principles.
Research ideas
In order to explore the role of LncRNA-Tug1 in the development of DN, this study used the US Arraystar RNA-seq platform to analyze the transcripts of glomeruli in type 2 diabetic mice and normal mice, and screen differentially expressed lncRNA. Among them, lncRNA-Tug1 was down-regulated in diabetic mice. Glucose stress experiments in in vitro podocytes and LOF and GOF in mice demonstrate that Tug1 is involved in the development of DN.
To explore the mechanism by which Tug1 regulates DN, the authors knocked down LncRNA-Tug1 in mouse podocytes. The expression profile chip analyzed the transcriptome and found 400 up-regulated genes associated with Tug1 and nearly 560 down-regulated genes. GO analysis revealed that Tug1 is involved in the regulation of several metabolic and biosynthetic pathways, and that the gene cluster associated with PCG-1α is significantly underexpressed in Tug1 knockdown mice. PCG-1α is a major regulator of mitochondrial function and is significantly under-represented in diabetic patients. Tug1 is associated with PCG-1 α and has become the focus of DN development. When excavating Tug1 and PCG-1 α, the authors first demonstrated that Tug1 affects mitochondrial bioenergy by regulating the expression of PCG-1α, and determines the target of Tug1, and focuses on the mechanism by which Tug1 regulates PCG-1α expression. Using motif analysis and CHIRP-seq, the authors found a TBE element in the upstream promoter region of Ppargc1 alpha that binds to Tug1. Promoter-luciferase and knockout experiments demonstrated that Tug1 interacts with the TBE element upstream of the Ppargc1α promoter to regulate PCG-1α expression. The expression of Ppargc1 α is self-regulating. The authors immediately demonstrated that Tug1 directly binds to the R/S domain of the CTD region of PCG-1α protein by RNA IP and Pull-down; CHIP experiments of PGC-1α demonstrated a large amount of TBE enrichment. Thus, it was revealed that the combination of Tug1 and TBE recruited PCG-1α protein, thereby realizing the expression regulation of Ppargc1α.
Technical route
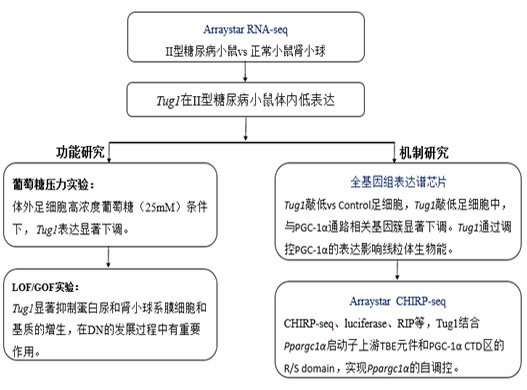
Result display
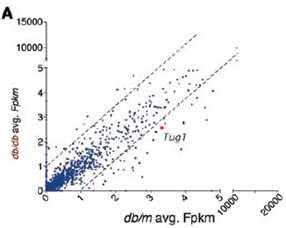
Figure 1 RNA-seq analysis differentially expressed lncRNA

Figure 2. Expression profile chip analysis, significant down-regulation of genes associated with PGC-1α pathway
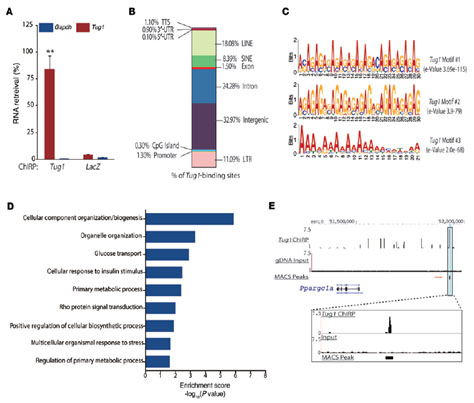
Figure 3. CHIRP-seq analysis reveals Tug1 binding site
Significance
This study revealed the mechanism by which LncRNA-Tug1 acts on mitochondrial bioenergy. LncRNA-Tug1 binds to the TBE element upstream of the Ppargc1α gene promoter, recruits PGC-1α protein, and binds to its CT/R domain R/S domain to achieve self-regulation of Ppargc1α gene, which in turn affects mitochondrial bioenergy. It provides a new idea for the exploration and treatment of DN pathology.
about the author
Farhad R. Danesh is a professor at the University of Texas at Anderson Cancer Center and a professor at Baylor College of Medicine. He has been engaged in research in the field of kidney disease for many years and has published many papers in the classic kidney disease journal JASN. In addition, more than 50 papers have been published in internationally renowned journals such as cell metabolism, Nature communication, and Kidney International. During the period, he also led the NIH/NIDDK support project, Mechanisms of protein-energy wasting in chronic kidney disease, Rho Kinases in diabetic nephropathy, which is second to none in the field of kidney disease research.
Original source
Long noncoding RNA Tug1 regulates mitochondrial bioenergetics in diabetic nephropathy. JCI.2016
Electrical Engineering,Winch Cable,Low Voltage Electrical Wire,High Voltage Electrical Wire
Shandong Freedoms Technology Co.,Ltd , https://www.sdfreedomtech.com